Blog
Estimation of Aberrant Cell Percentage in Tumor Normal Cell Mixture
Cancer samples, especially solid tumors, usually contain a mixture of tumor and normal cells. Using both the SNP and copy number information provided by SNP arrays (Affymetrix or Illumina), the % aberrant cells can be estimated. Here we are going to discuss a simple formula for such calculation based on one-copy loss event.
Figure below shows the B-Allele Frequency (BAF) bands in a region where a mixture of aberrant cells with one-copy loss and non-aberrant/normal cells is present.
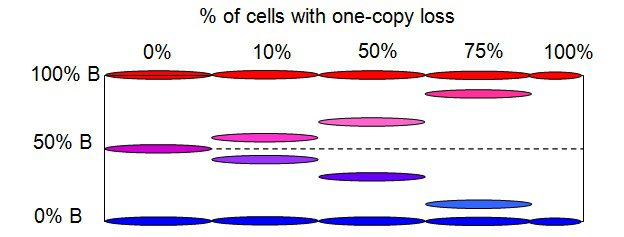
With no aberrant cells (0%), or only normal cells, a typical 3-band BAF pattern is displayed on the left, indicating AA (0% BAF), AB (50% BAF), and BB (100% BAF) alleles. With only aberrant cells (100%) of one-copy loss, a typical 2-band BAF pattern is shown on the right, a representation of either A (0% BAF) or B (100% BAF) allele. When there is a mixture of these two types of cells, two middle BAF bands will appear. And the location of the two middle bands is dependent upon the % of aberrant cells.
Because the two middle bands are mirror images of each other around 50% BAF, only the lower band is needed for the calculation of the % aberrant cells. A simple formula is deduced for such calculation:
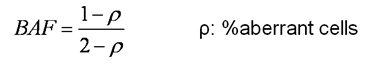
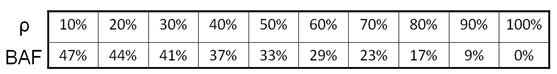
And based on this formula, different % aberrant cells and their related BAF values are listed in this table:
Interested in learning more? View our on-demand webinar, “Combined Analysis and Interpretation of CNV, AOH, and Seq Var of FFPE Cancer Samples from a Solid Tumor”
